Using Scripts¶
We use scripts to use the functionalities offered by cytocalc. Scripts provide an interface for using cytocalc while maintaining a relatively
stable ‘core’ functions. System specific information (file structure, folder names etc.)
are handled using the scripts, which hence change more often than code in the src
folder (eg: function for computing network radius) which doesn’t have to be modified a lot.
The scripts directory provides a few pre-made scripts for some common systems.
These are intended to be used as a starting point for creating more complex scripts.
Example¶
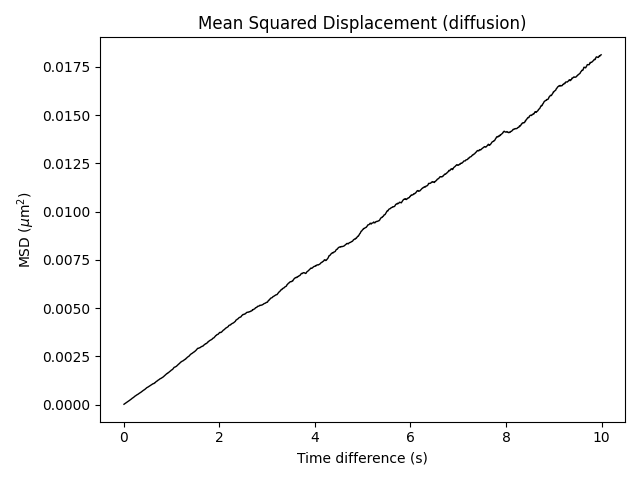
Suppose we would like to measure the Mean Squared Displacement of brownian particles
using the diffusing_bead_msd.py script. The simulation details which the script has
been designed for are as follows:
Initialize N beads, non interacting
Run the simulation for T time
Use
reportFto produce report files in<directory>
To run the script, we must first ensure that the cytocalc is available in the current
python environment. This can be done by switching to the virtual environment created during
Installation.
$ source $CYTOCALCPATH/.venv/bin/activate
where $CYTOCALCPATH is the path to the local Cytocalc repository. We can now run the script
with the directory location as the argument.
(.venv) $ $CYTOCALCPATH/scripts/diffusing_bead_msd.py <directory>